2YPZ
 
 | |
4UZB
 
 | | KSHV LANA (ORF73) C-terminal domain mutant bound to LBS1 DNA (R1039Q, R1040Q, K1055E, K1109A, D1110A, A1121E, K1138S, K1140D, K1141D) | | Descriptor: | LANA BINDING SITE 1 DNA, ORF 73 | | Authors: | Hellert, J, Krausze, J, Luhrs, T. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.865 Å) | | Cite: | The 3D Structure of Kaposi Sarcoma Herpesvirus Lana C-Terminal Domain Bound to DNA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A76
 
 | |
4K2J
 
 | |
4UZC
 
 | |
7BCY
 
 | |
2YQ0
 
 | |
2YPY
 
 | |
7BED
 
 | |
2YQ1
 
 | |
1ZLA
 
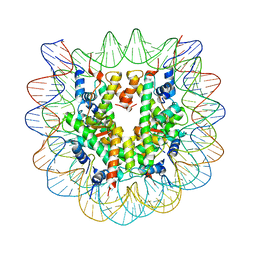 | | X-ray Structure of a Kaposi's sarcoma herpesvirus LANA peptide bound to the nucleosomal core | | Descriptor: | Histone H4, Palindromic 146bp Human alpha-Satellite DNA fragment, Xenopus laevis-like histone H2A, ... | | Authors: | Chodaparambil, J.V, Barbera, A.J, Kaye, K.M, Luger, K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA.
Science, 311, 2006
|
|
6VH6
 
 | |
6LYV
 
 | | The crystal structure of SAUGI/KSHVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
4PLX
 
 | | Crystal structure of the triple-helical stability element at the 3' end of MALAT1 | | Descriptor: | Core ENE hairpin and A-rich tract from MALAT1 | | Authors: | Brown, J.A, Bulkley, D, Wang, J, Valenstein, M.L, Yario, T.A, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6LYJ
 
 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
